Enzyme Genetics
Introduction
In biological research, there are several occasions in which genetic comparisons are needed. For instance, there are times when there is not much information on a specific organism or enzyme that is of interest to the researcher. Therefore it is useful to be able to find genetically similar organisms or enzymes that are better researched. The genetics can help justify the use of these organisms as a base for setting up an experiment or background for an experiment. Additionally, genetics can be used to determine the evolutionary relationships between organisms. The more genetically similar organisms or enzymes are to each other then the more recently they branched from a common ancestor. It is therefore useful to become familiar with this type of software.
National Center for Biotechnology Information (NCBI) is a database where, among other things, genes can be easily searched for and found. Basic Local Alignment Search Tool (BLAST) is a tool on NCBI that allows the comparison of a query sequence with all other sequences in the database.
Concepts
The subject for this project to familiarize people with the database and tools is the enzyme Glutathione S-Transferase (GST) in Mus musculus (house mouse). The mouse was chosen because its genome has been extensively researched and sequenced. GSTs are enzymes that have been found to be a part of the cell’s protection from multiple dangerous chemicals that are produced both in the environment and within the organism.
GSTs are split into three different superfamilies. The largest of these superfamilies is the cytosolic and this superfamily has thirteen recognized classes: alpha, delta, beta, epsilon, zeta, theta, mu, nu, pi, sigma, tau, phi and omega. You can recognize each superfamily by the letter that directly follows Gst in the name section (i.e. the alpha superfamily is Gsta) and then each superfamily has different categories under them, which are distinguished by numbers.
Conserved Domains are sections of the genome that are similar in several different species. This suggests that they perform an important function for the organism and therefore it has been preserved through evolution.
Supplies
Computer
Internet connection
Websites
http://www.ncbi.nlm.nih.gov
http://blast.ncbi.nlm.nih.gov/Blast.cgi
Time
This project should not take more than 20 minutes.
In biological research, there are several occasions in which genetic comparisons are needed. For instance, there are times when there is not much information on a specific organism or enzyme that is of interest to the researcher. Therefore it is useful to be able to find genetically similar organisms or enzymes that are better researched. The genetics can help justify the use of these organisms as a base for setting up an experiment or background for an experiment. Additionally, genetics can be used to determine the evolutionary relationships between organisms. The more genetically similar organisms or enzymes are to each other then the more recently they branched from a common ancestor. It is therefore useful to become familiar with this type of software.
National Center for Biotechnology Information (NCBI) is a database where, among other things, genes can be easily searched for and found. Basic Local Alignment Search Tool (BLAST) is a tool on NCBI that allows the comparison of a query sequence with all other sequences in the database.
Concepts
The subject for this project to familiarize people with the database and tools is the enzyme Glutathione S-Transferase (GST) in Mus musculus (house mouse). The mouse was chosen because its genome has been extensively researched and sequenced. GSTs are enzymes that have been found to be a part of the cell’s protection from multiple dangerous chemicals that are produced both in the environment and within the organism.
GSTs are split into three different superfamilies. The largest of these superfamilies is the cytosolic and this superfamily has thirteen recognized classes: alpha, delta, beta, epsilon, zeta, theta, mu, nu, pi, sigma, tau, phi and omega. You can recognize each superfamily by the letter that directly follows Gst in the name section (i.e. the alpha superfamily is Gsta) and then each superfamily has different categories under them, which are distinguished by numbers.
Conserved Domains are sections of the genome that are similar in several different species. This suggests that they perform an important function for the organism and therefore it has been preserved through evolution.
Supplies
Computer
Internet connection
Websites
http://www.ncbi.nlm.nih.gov
http://blast.ncbi.nlm.nih.gov/Blast.cgi
Time
This project should not take more than 20 minutes.
Go to the National Center for Biotechnology Information
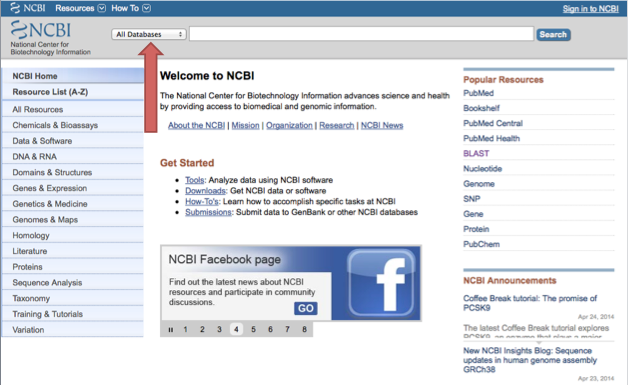
Go to http://www.ncbi.nlm.nih.gov
Then change "All Databases" to "Gene" in the box next to the search bar.
Then change "All Databases" to "Gene" in the box next to the search bar.
Search the Database
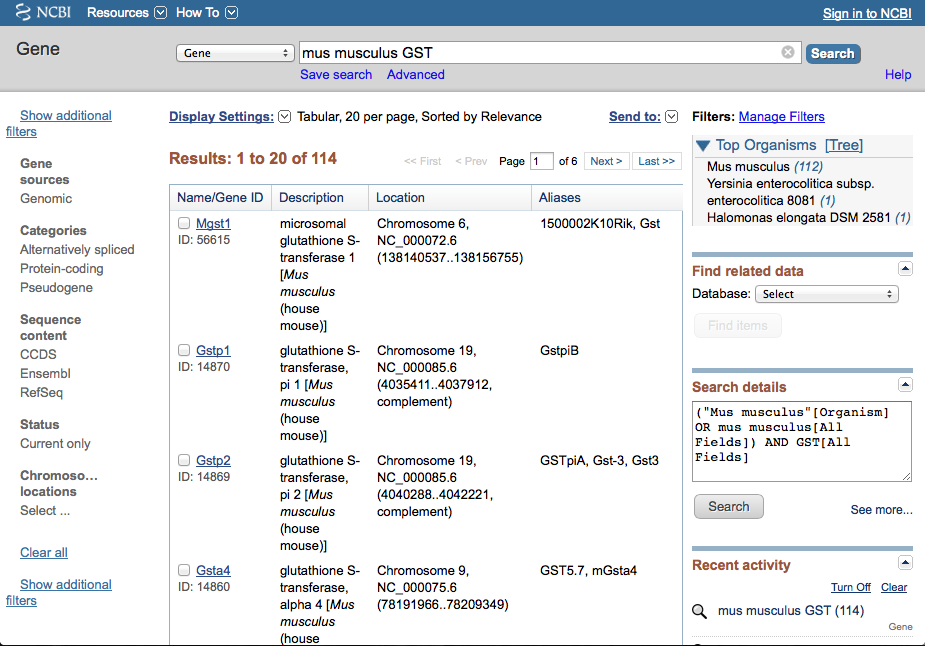
Type "Mus musculus GST" into the search bar and click on search.
Pick a Specific Gene

Since the purpose of this instructable is to become familiar with the software, it does not matter which superfamily or category you choose.
The gene used for the duration of the instructions and is therefore in all of the pictures is Gsta1. Just click on the name of the gene (in purple in the picture but it will be blue on the website) that you choose to get to the next step.
The gene used for the duration of the instructions and is therefore in all of the pictures is Gsta1. Just click on the name of the gene (in purple in the picture but it will be blue on the website) that you choose to get to the next step.
Get the Sequence
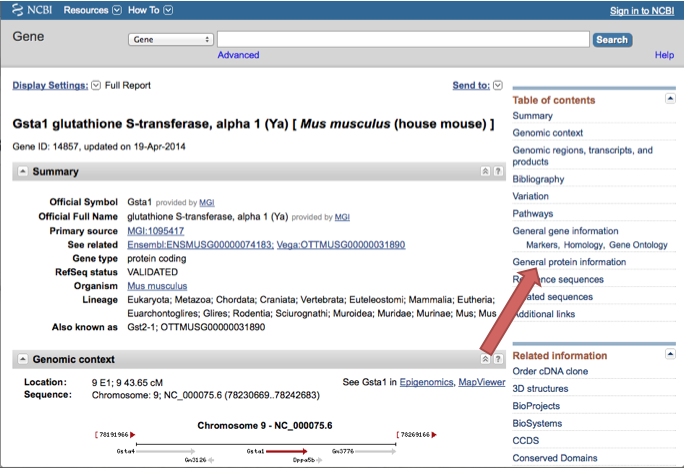
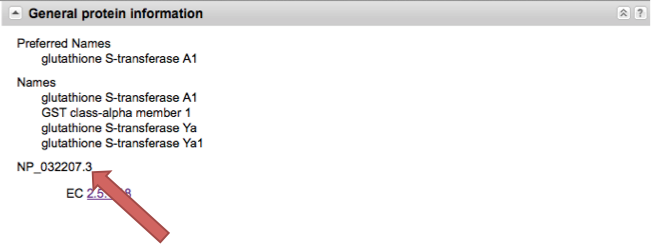
Click on General Protein Information on the right side of the screen.
Copy the accession number (this should start with NP_ and is followed by a string of numbers). When you copy this, make sure the NP_ is included.
Copy the accession number (this should start with NP_ and is followed by a string of numbers). When you copy this, make sure the NP_ is included.
Go to BLAST
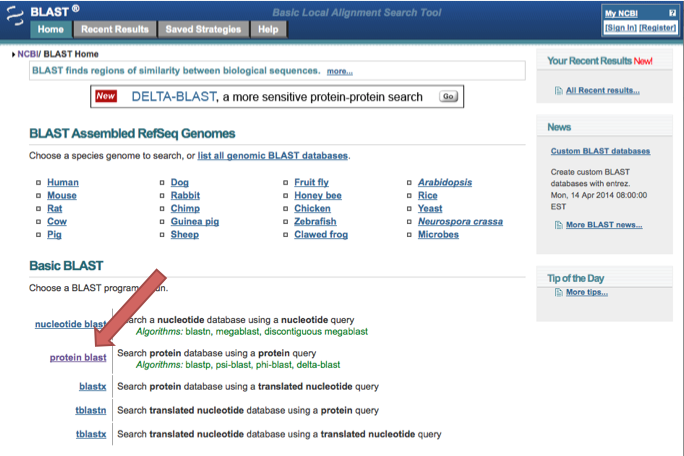
Go to http://blast.ncbi.nlm.nih.gov/Blast.cgi and click on "protein blast"
Enter Query
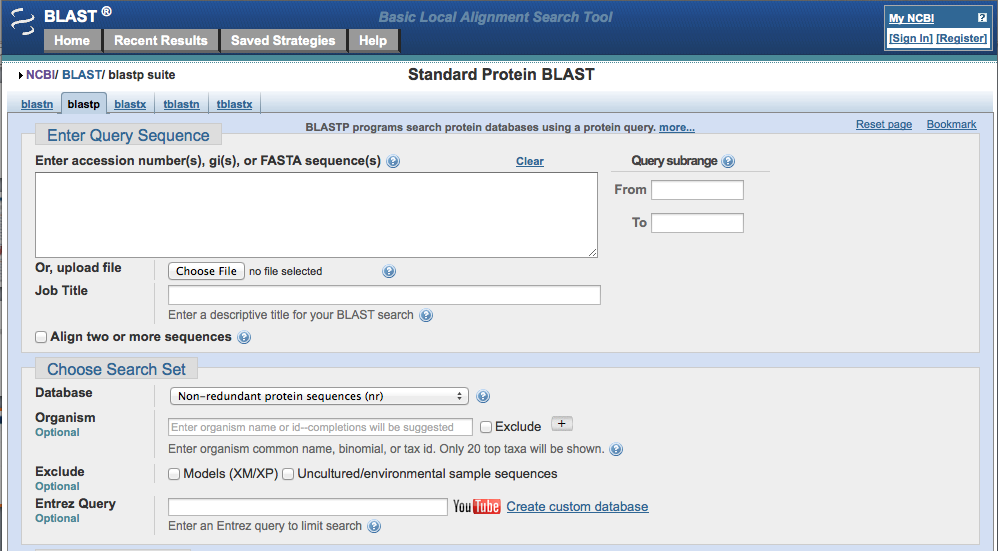
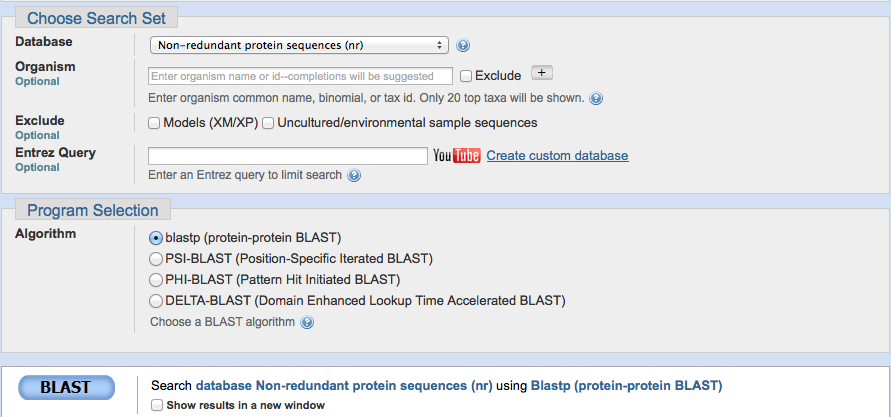
Paste the accession number.
Make sure that the database is on "non-redundant protein sequences (nr)" and the Algorithm is "blast (protein-protein BLAST)." You don't have to worry about any of the other sections or boxes.
Click the BLAST button at the bottom of the page.
Make sure that the database is on "non-redundant protein sequences (nr)" and the Algorithm is "blast (protein-protein BLAST)." You don't have to worry about any of the other sections or boxes.
Click the BLAST button at the bottom of the page.
Results Page
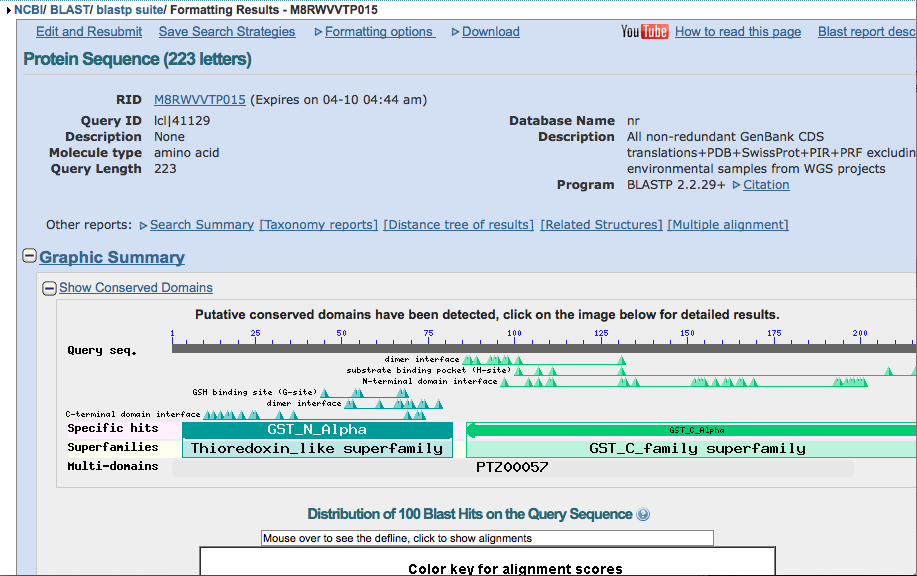
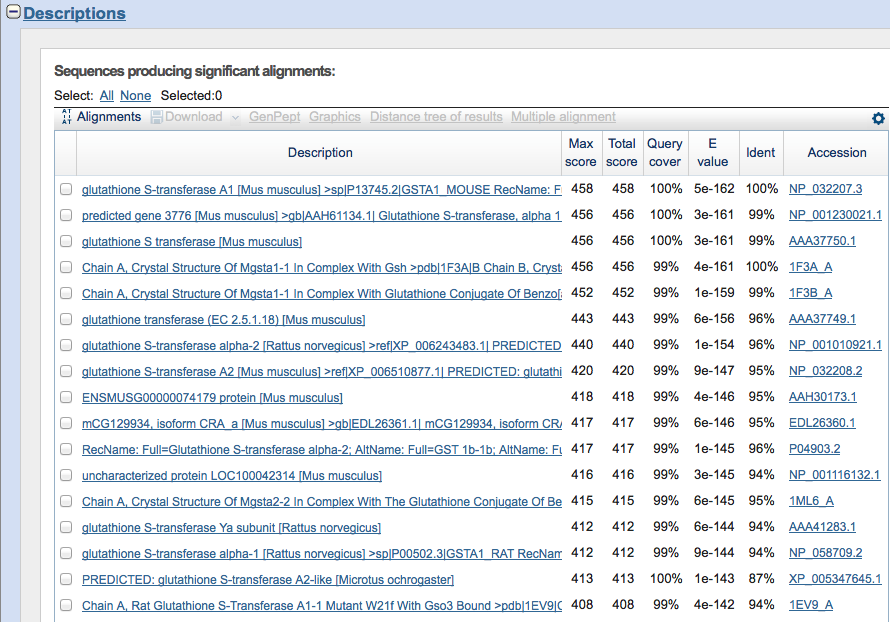
There should be several redirects, the same page might reload several times, and it could take some time to get to the results page. The first picture is of the page that you will see when you are waiting for the results.
Once on the results page scroll down to the "descriptions" section.
Once on the results page scroll down to the "descriptions" section.
Select Sequences to Compare
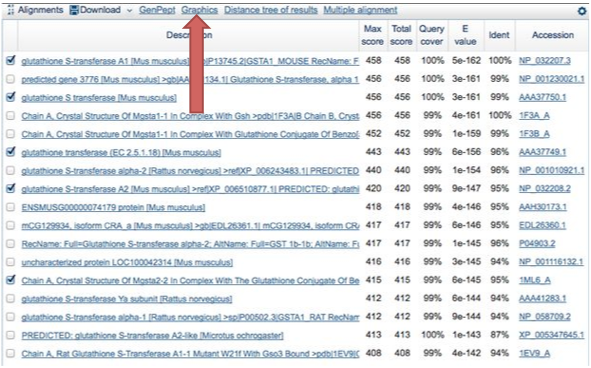
The sequences in the "Description" section are organized by how similar they are to the query.
Click the boxes next to the sequences that you want to compare side-by-side. Again, since the purpose is to familiarize yourself with the software, It does not matter which ones you choose.
Then click on the "Graphics" link on the top of the descriptions box.
Click the boxes next to the sequences that you want to compare side-by-side. Again, since the purpose is to familiarize yourself with the software, It does not matter which ones you choose.
Then click on the "Graphics" link on the top of the descriptions box.
Find Conserved Domains
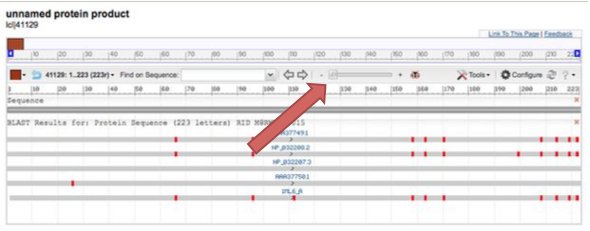
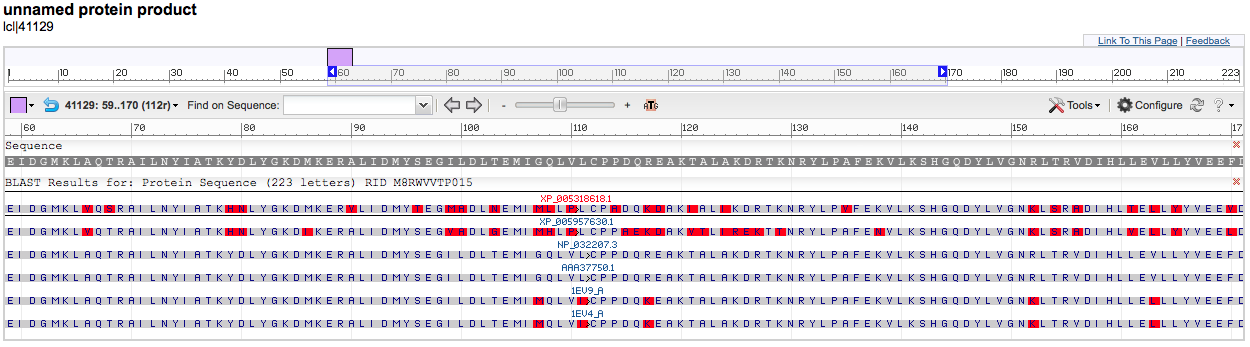
The red bars are the conserved domains in the protein sequence.
You can zoom in and out to see the actual sequence and move the view up and down the protein. This is helpful in finding the conserved domains across the different species.
You can zoom in and out to see the actual sequence and move the view up and down the protein. This is helpful in finding the conserved domains across the different species.
Comparing Two Sequences
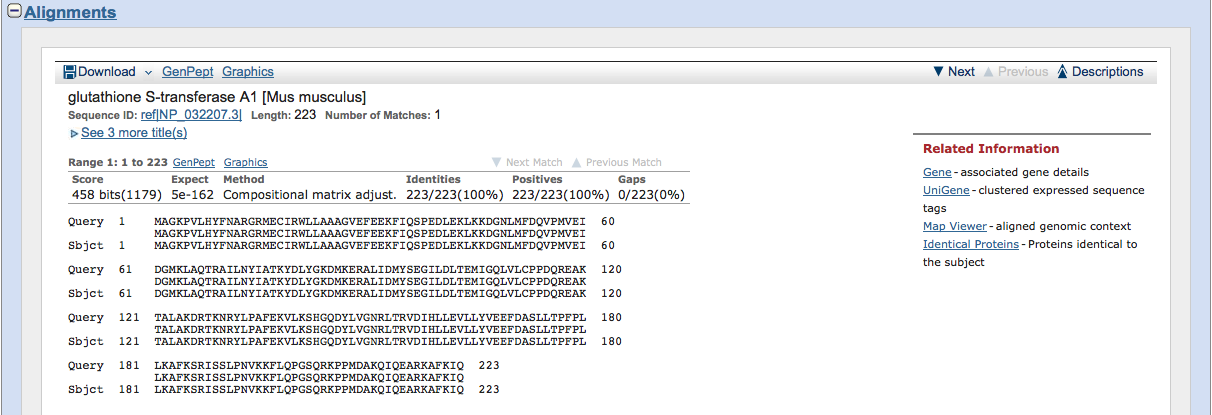
If you only want to compare the sequences of the query and another protein from the "Descriptions" then you can click on one from the list and go to the "Alignments" section.
This lines up the query (accession number you typed in) and the subject (gene you clicked on in "Descriptions"), with the similar proteins listed in the middle line. If there is not a protein in common, then there will be a blank space.
You are now familiar with the basic functions of these websites if you ever need to use genetic comparisons in research.
This lines up the query (accession number you typed in) and the subject (gene you clicked on in "Descriptions"), with the similar proteins listed in the middle line. If there is not a protein in common, then there will be a blank space.
You are now familiar with the basic functions of these websites if you ever need to use genetic comparisons in research.