3D Print a Molecular Surface: L-A Virus Capsid
by RNAlab in Workshop > 3D Printing
365 Views, 4 Favorites, 0 Comments
3D Print a Molecular Surface: L-A Virus Capsid




Bring the microscopic world of proteins to the macro scale. With this tutorial you can convert any protein surface into a 3D printed object. Before you would be limited to structures with solved crystal structures, but now with AI "solving" protein folding, almost any structured protein is a click away.
Project preamble: L-A virus infects almost all laboratory strains of Baker's Yeast (Saccharomyces cerevisia) and many wild strains. Once infected, yeast pass the virus down from mother to daughter. Even though it was discovered over 40 years ago, it is still unknown how the virus transmits from one infected yeast cell to another.
While not known to cause overt phenotypes itself, another virus called M Satellite can super-infect the yeast and hijack the molecular machinery of L-A virus, including it's capsid. M Satellite encodes for a Killer Toxin system which kills neighbouring yeast cells. When brewing beer, M Satellite infected yeast can contaminate yeast cultures, and kill the tasty-fermenting strains, causing bad brew (spoilage).
Supplies

Software
- PyMol Protein Viewer (Trail/Educational) or PyMol Open Source (requires compile)
- Blender (Open Source)
- Atomic Blender Add-on (OPTIONAL, imports protein .pdb file)
- PrusaSlicer (or your favourite slicer software)
Hardware
- 3D printer. I used a MK4.
- 3D print filament (variable). Silk duotone PLA looks nice. 300g
- 3D print support clean-up supplies (flush cutters, exacto, pliers)
Print Files
- L-A Virus (1M1C) Example Print Example
- Gamma Secretase + Beta-Amyloid (Alzeimer's Protein) Print Example
Choose your own protein:
Download an Atomic-resolution Model of Your Virus From PDB

The PDB Database (https://www.rcsb.org) houses crystal and electron-microscope structures for proteins, including many “Virus Assembly” models. Use the advanced search to choose an available virus particle. We will select L-A Virus (pdb: 1M1C). Click Download and select Biological Assembly 1 (CIF -gz) format. The .pdb or .cif formats are the standard formats for storing atomic protein structures.
Load the Model Into PyMol

PyMol (https://github.com/schrodinger/pymol-open-source) is a protein visualization tool. Install PyMol and load the `cif.gz` biological assembly file. This will give you a wire-diagram of the backbone carbon ribbon of the protein.
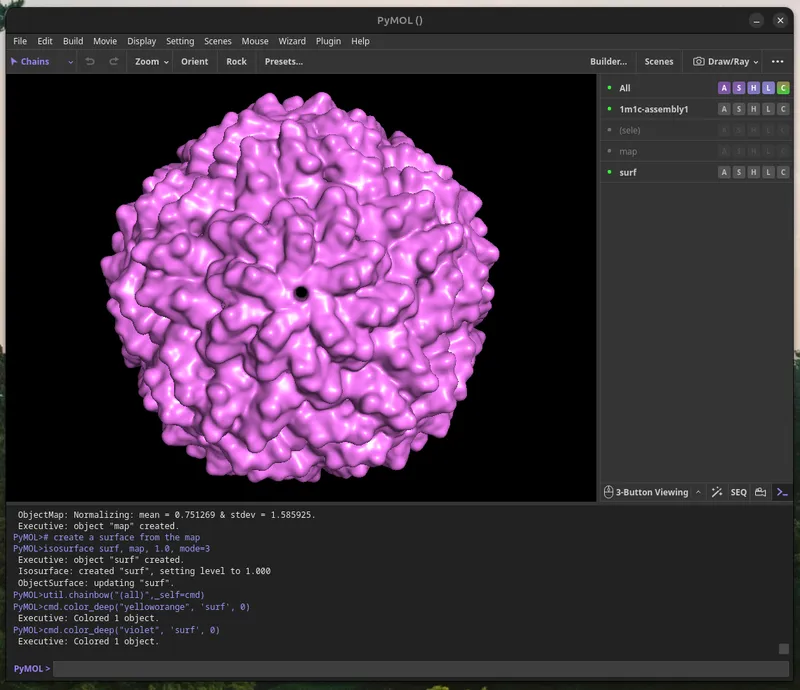
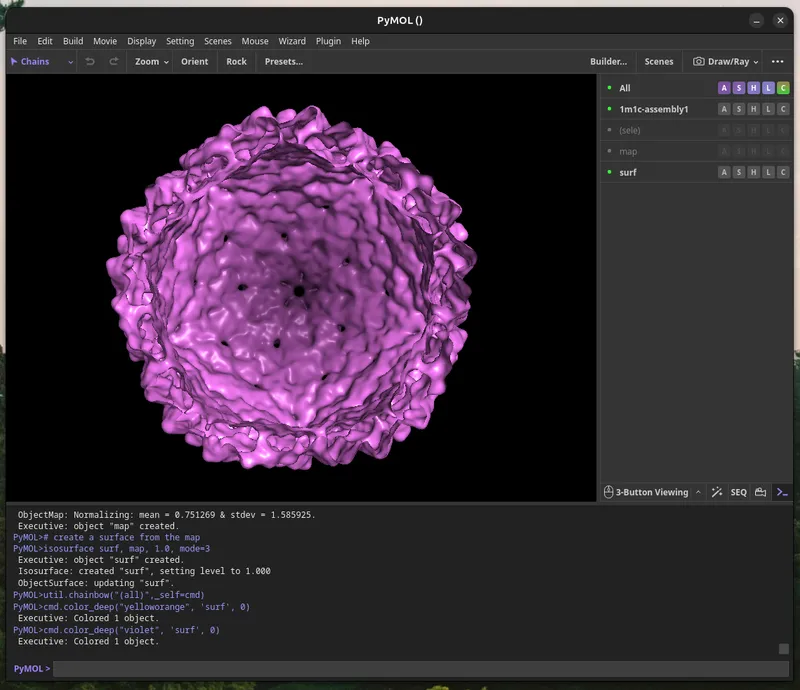
The L-A Virus particle is made up of 120 copies of one protein, called the capsid. These spontaneously form an icosahedral (20-sided), inside of which the double-stranded RNA virus genome is packaged, and protected from cellular protein attack. Where the 5 protein edges (magenta) intersect at a vertex, L-A virus has a pore which is used to shoot out viral messenger RNA into infected cells to make more virus capid proteins.
Calculate the Molecular Surface for the Assembly
You will calculate a “Molecular Surface” for your model, this can take a few minutes. Paste the commands below into PyMol for a basic setting I found gives aesthetic prints. Enjoy a gummy while you wait (2 minutes - several hours, depending on resolution).
For more information about these parameters for large surface rendering, see PyMol Wiki for Huge Surfaces.
Export the Molecular Surface As .wRL
When you're satisfied with your molecular surface map, once it has the right level of detail, you can export it using `File > Save Image As > VRML2` and save the surface.
Load the WRL Into Blender


To prepare the model for a high-quality print, I use Blender (https://www.blender.org/) to convert the `wrl` file into a clean `stl` file. Import the file you saved from PyMol using the `File > Import > X3D`
Optimize the Model for Printing
It's quite fun to play with Blender at this point for each model and figure out how to get a more aesthetic print made. Be creative, and most importantly have fun!
A few of essential pre-processing steps:
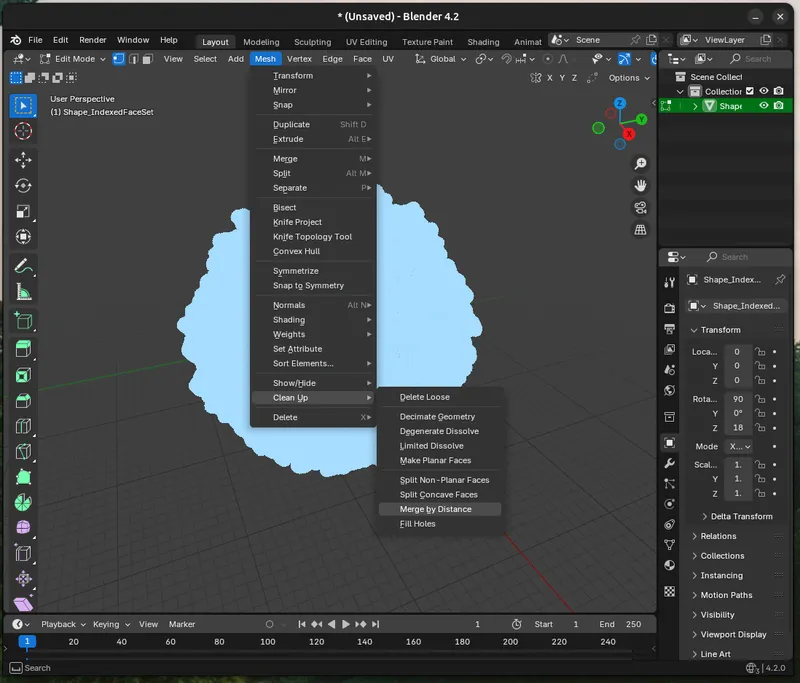
a. Merge By Distance to link the faces together, this makes your object “one thing" for many filters in Blender. Go to Edit Mode then select all (A), and click `Mesh > Clean Up > Merge By Distance`. Use `0.00001m` as the setting.
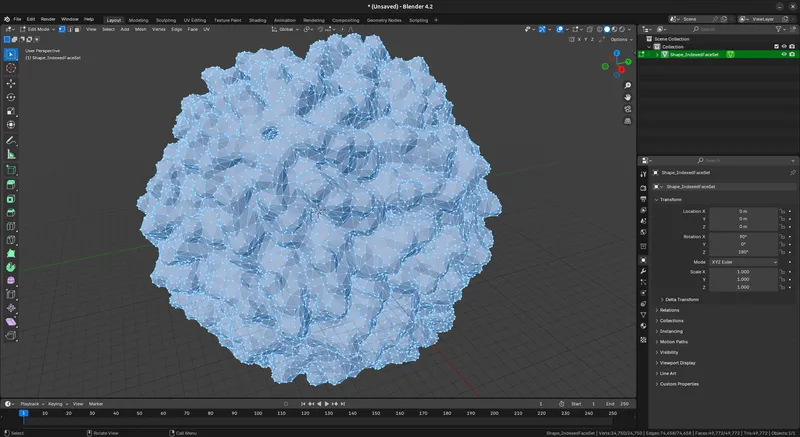
b. Decimate Geometry (Optional). I'm going to print with a glossy filament, to give each surface a distinct sheen, I want a more “polygon-ized” version of the model, with sharp edges to create facets. Use `Mesh > Clean Up > Decimate Geometry`. I use a value of `0.1` but varying this will change the effect. Keep in mind the value you set here and how it looks depends on the resolution parameters you used in PyMol For the L-A virus model, I reduce the original 248,732 vertices to 24,750 vertices.
c. Fill the Cavity (Optional). Depending on your virus/protein, you may want to (have to) fill the interior space of the model. I use a sphere with a centre coordinate of 0,0,0 and tweak the size in the text editor until it about fills the virus.
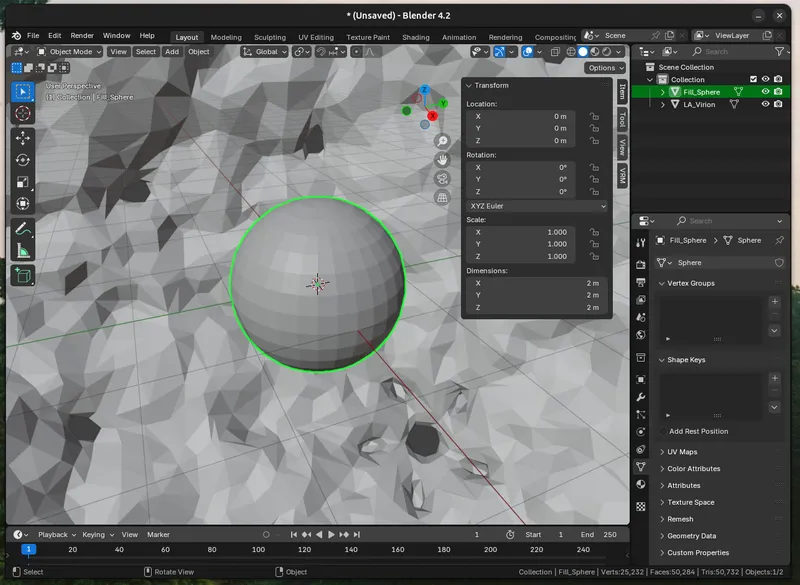
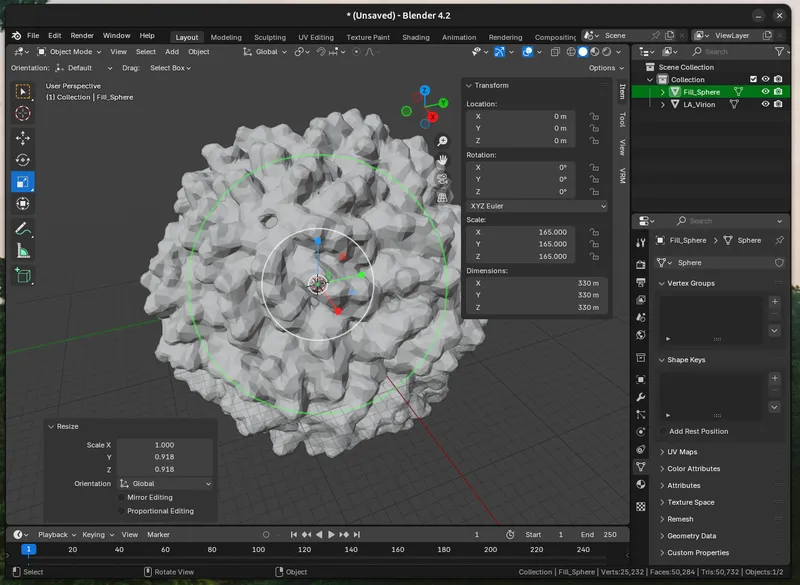
Once it's the right size to fill the interior, merge the two objects by creating a `Boolean Modifer ("Union")` modifier on the virion and sphere. `Add Modifier > Generate > Boolean`
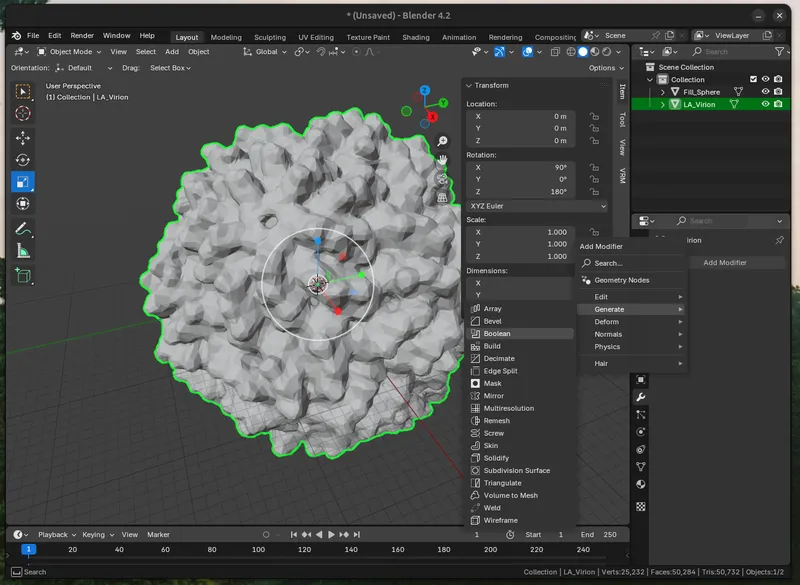
Then select the interior “Fill_Sphere” as the intersect object.
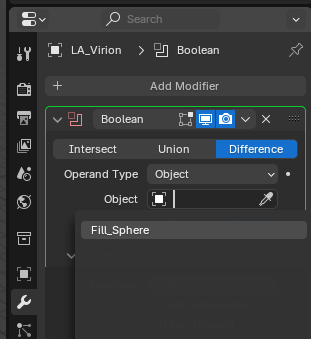
Once the Boolean fill looks good, and there are no weird overlaps or mis-calculations (it will happen). Apply the modifiers to match the `“Visual Geometry” (what you see). Object > Apply > Visual Geometry to Mesh`.
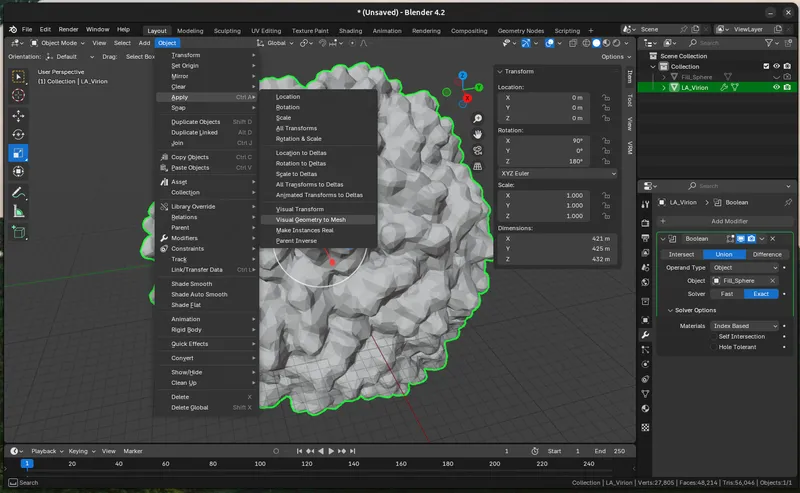
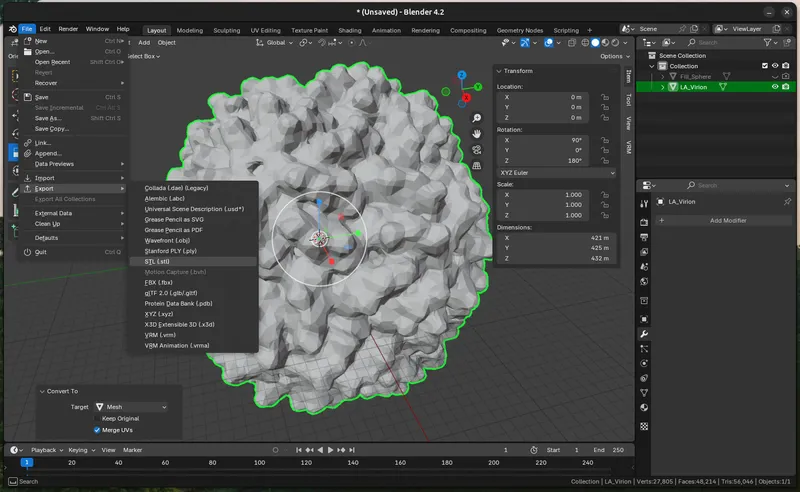
Export the Model (STL Conversion)
Save your .blender file. To export the model into a file-format recognized you 3D printer slicers, you'll need to export it as .stl: `File > Export > STL`
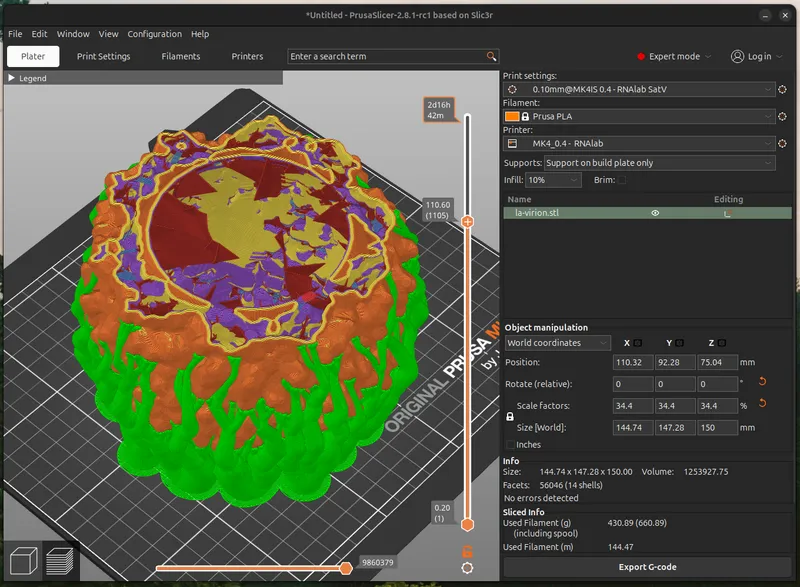
Model Slicing (gcode Conversion)
I use PrusaSlicer, to convert the 3D model into 3D printer instructions. Import your stl file (drag and drop), rotate it it if necessary to select a flat print surface.
There's no single correct choice for 3-D printing or slicing parameters. Many factors may influence practical and aesthetic considerations of your print. For a hand-held sized virion ( height: 150mm), printed with regular filament (material: Silk PLA dual-color filament) I found the following settings helpful
Nozzle Size: 0.4mm
Layer Height: 0.1mm
Perimeters: 4
Thick Bridges: True
Supports: Organic
Infill: Lightning, 10%
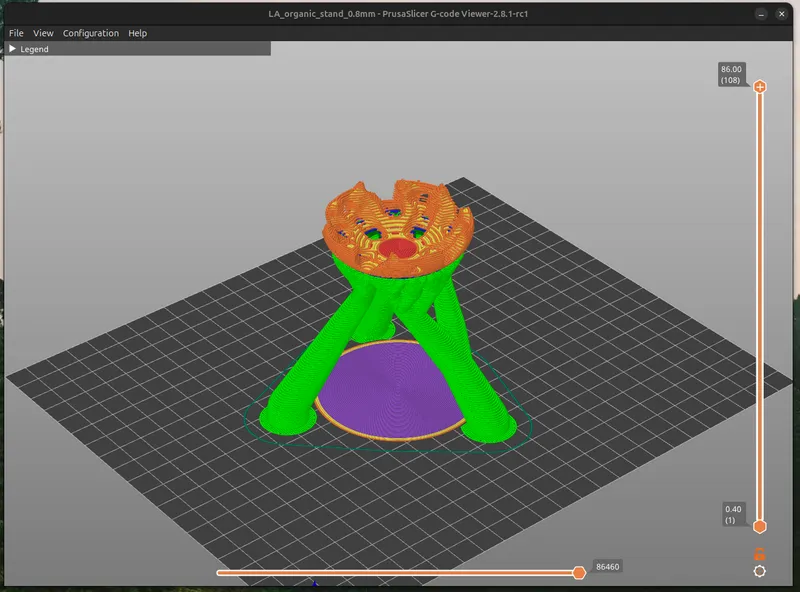
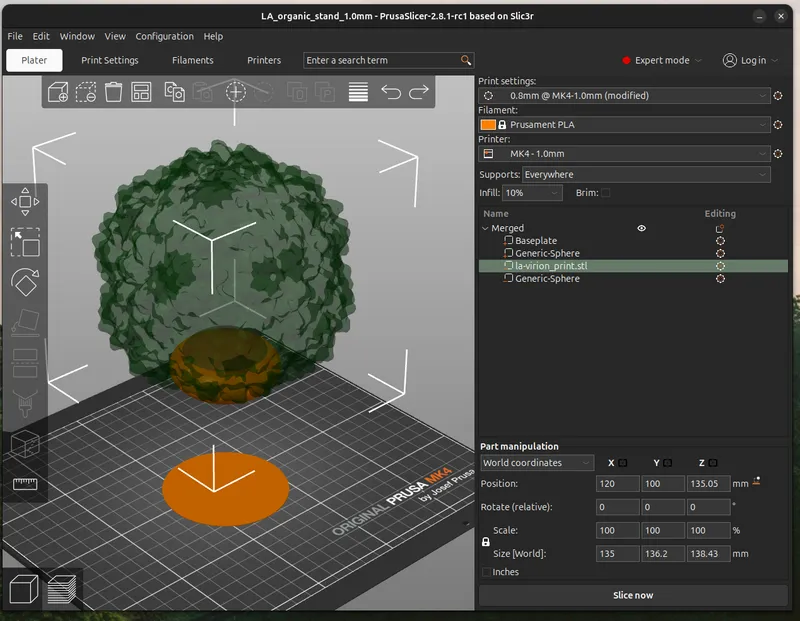
Print a Virus Stand
Anything can be made into a virus stand with this method, be creative.
I'm playing around with a cool organic style stand for the virion. Recently I've also been playing with a 1.0mm nozzle. Combining these elements, I use elevated shapes, with incuts of the virus as the contact points. The floating polygons get tree supports going up to them as a type of Support. I also add a 1-layer thick object (purple cylinder) to exclude the default downwards support legs and give the stand a steady base. Altogether this is aesthetically a good match for the natural shape of the virus.
I'm including the PrusaSlicer (3mf) project file for you to play around with to get a sense of how it works, consider it experimental.
3D Print + Share!

"happiness is only real when shared" -AS (CM)
If you got this far, show me your project I'd love to see which protein/virus you tackled.